20 changed files with 38 additions and 4 deletions
BIN
Results/.DS_Store
BIN
Results/gen_iter.png
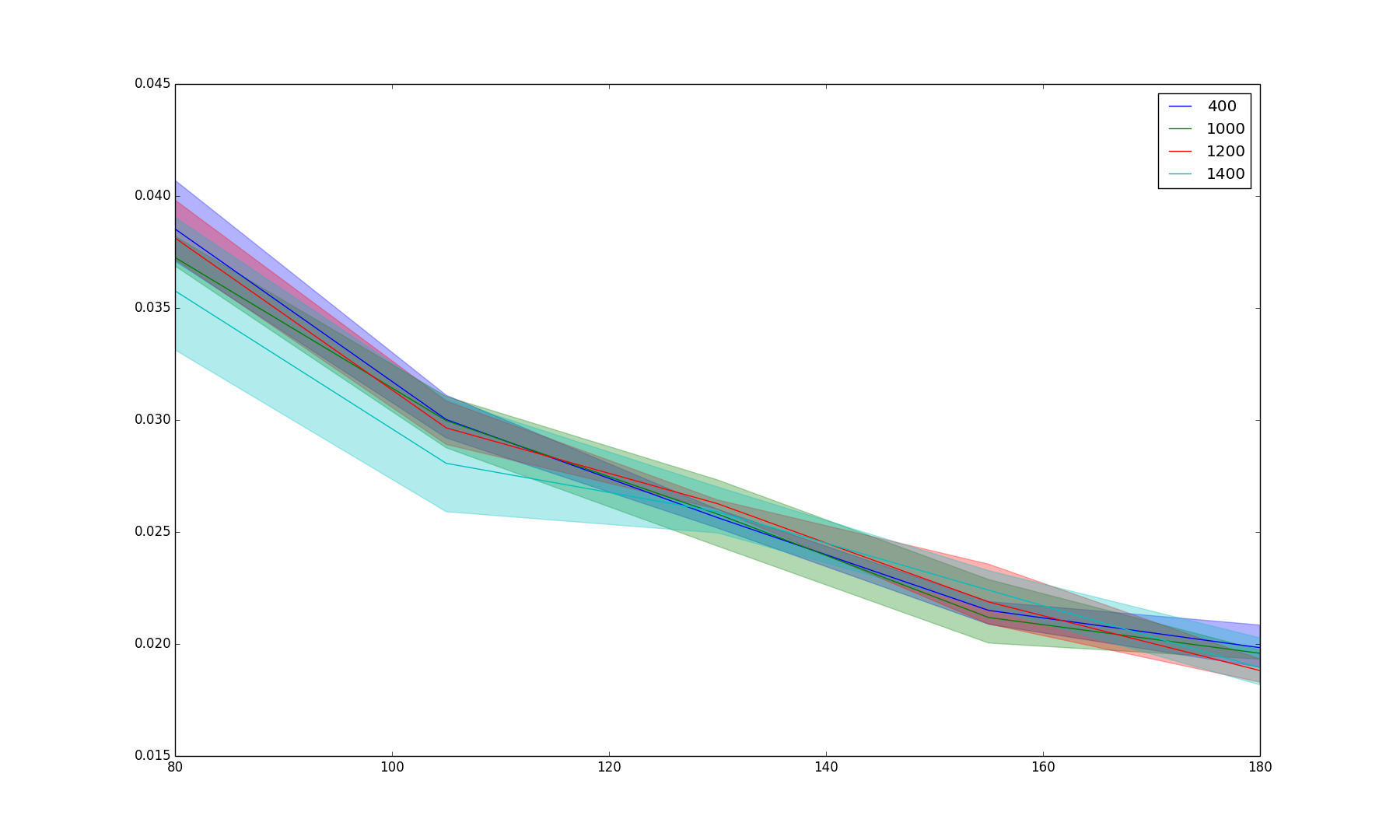
BIN
Results/random_iter.png
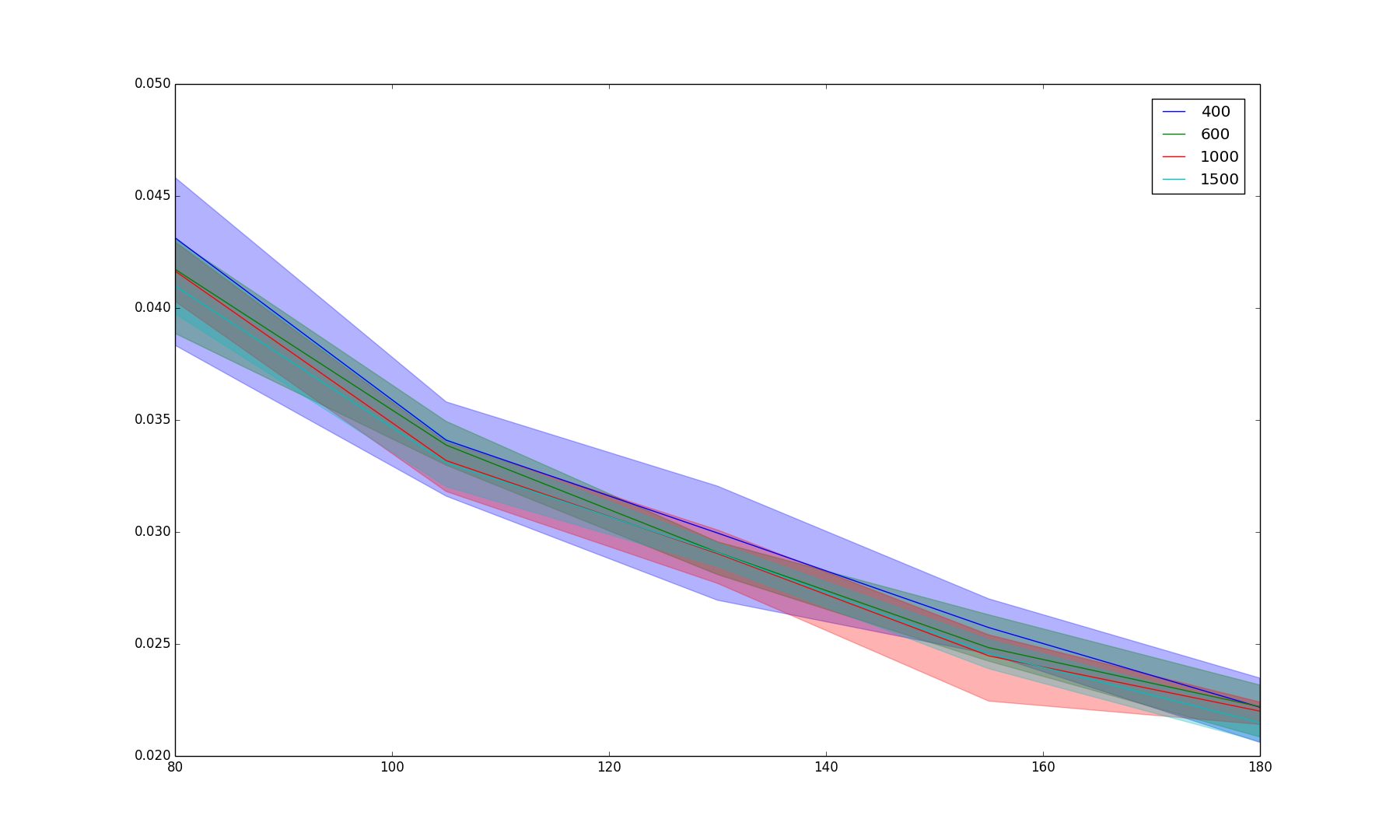
BIN
Results/random_iter_3.png
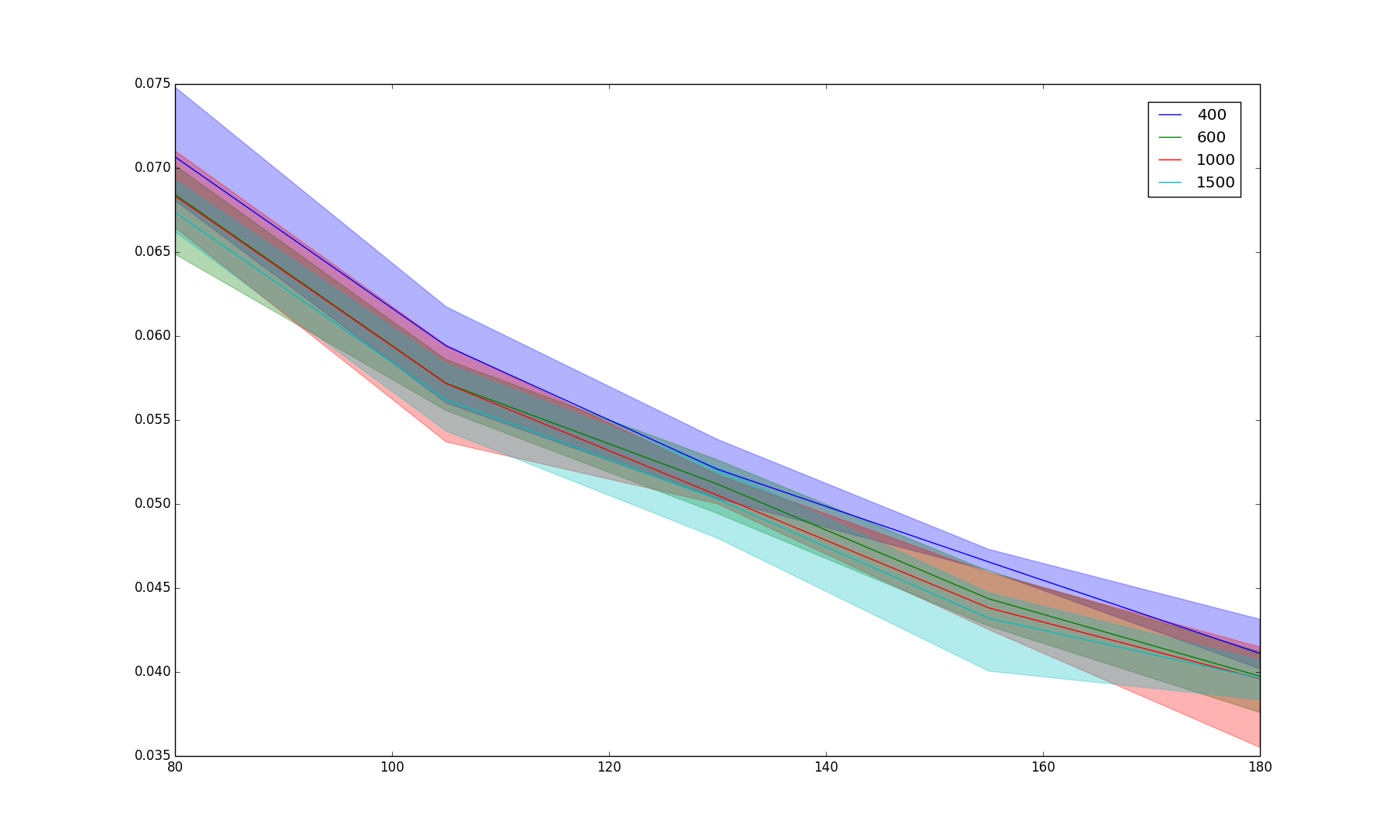
BIN
Results/res_gen_2.png
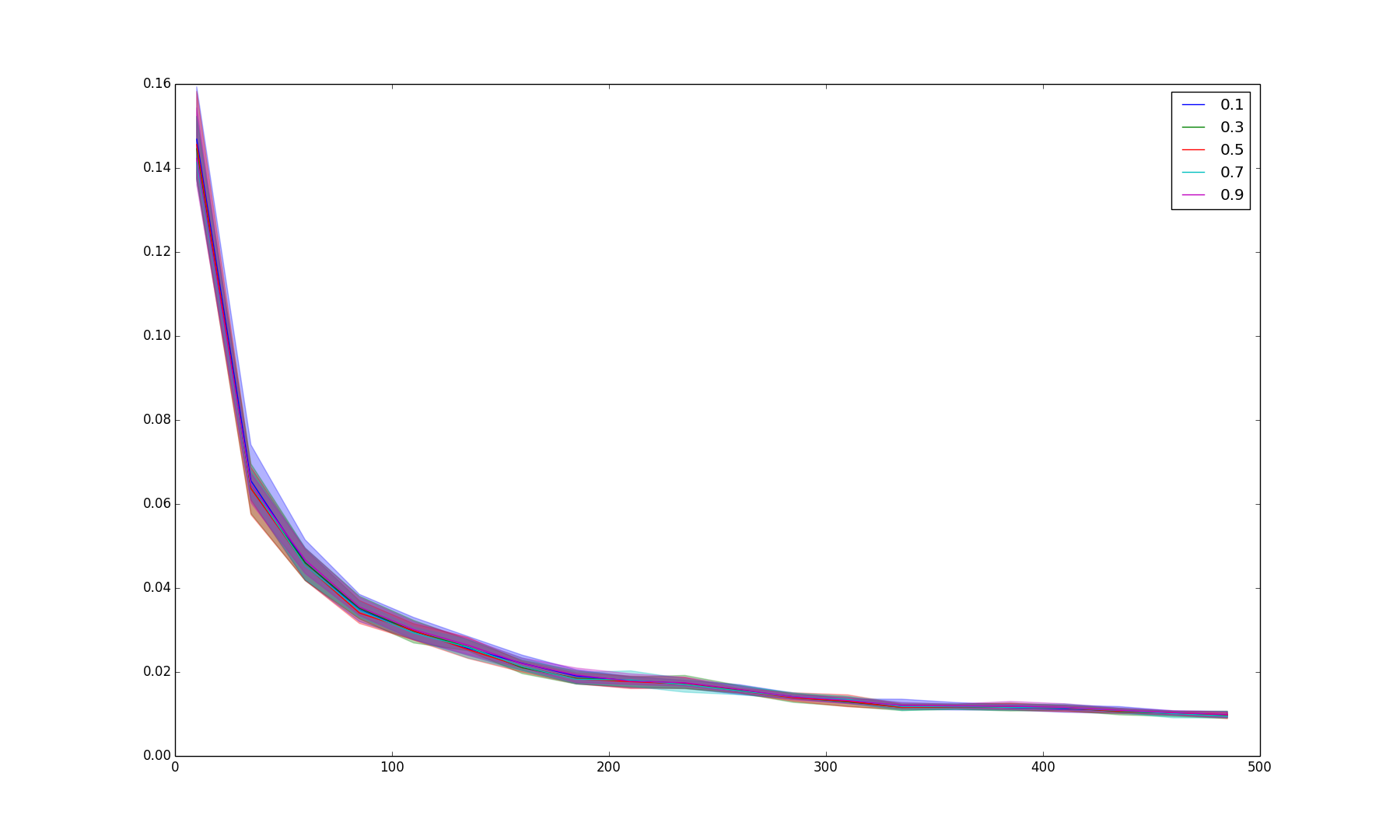
BIN
Results/res_gen_2_zoom.png

BIN
Results/res_gen_3_zoom.png
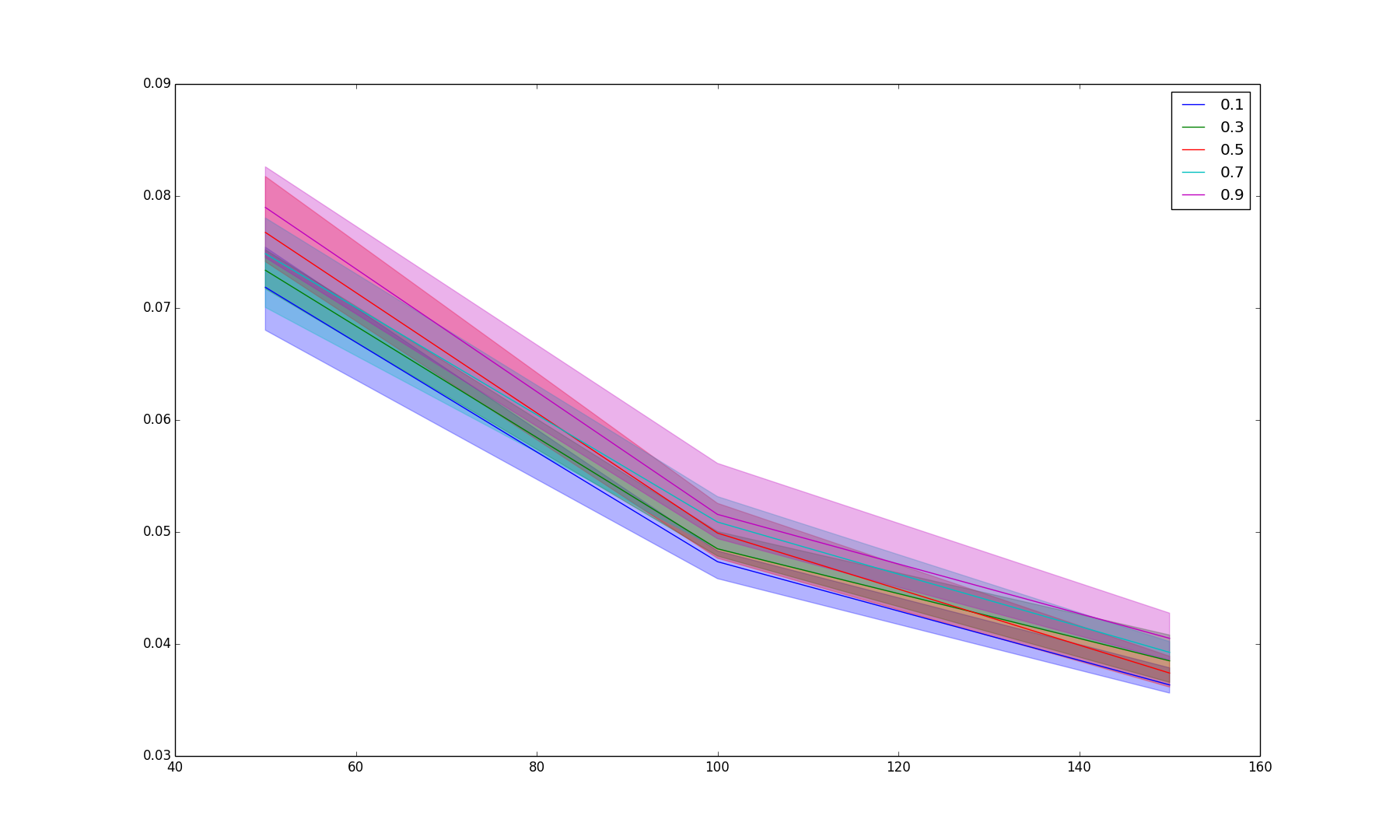
BIN
Results/res_gen_4_zoom.png
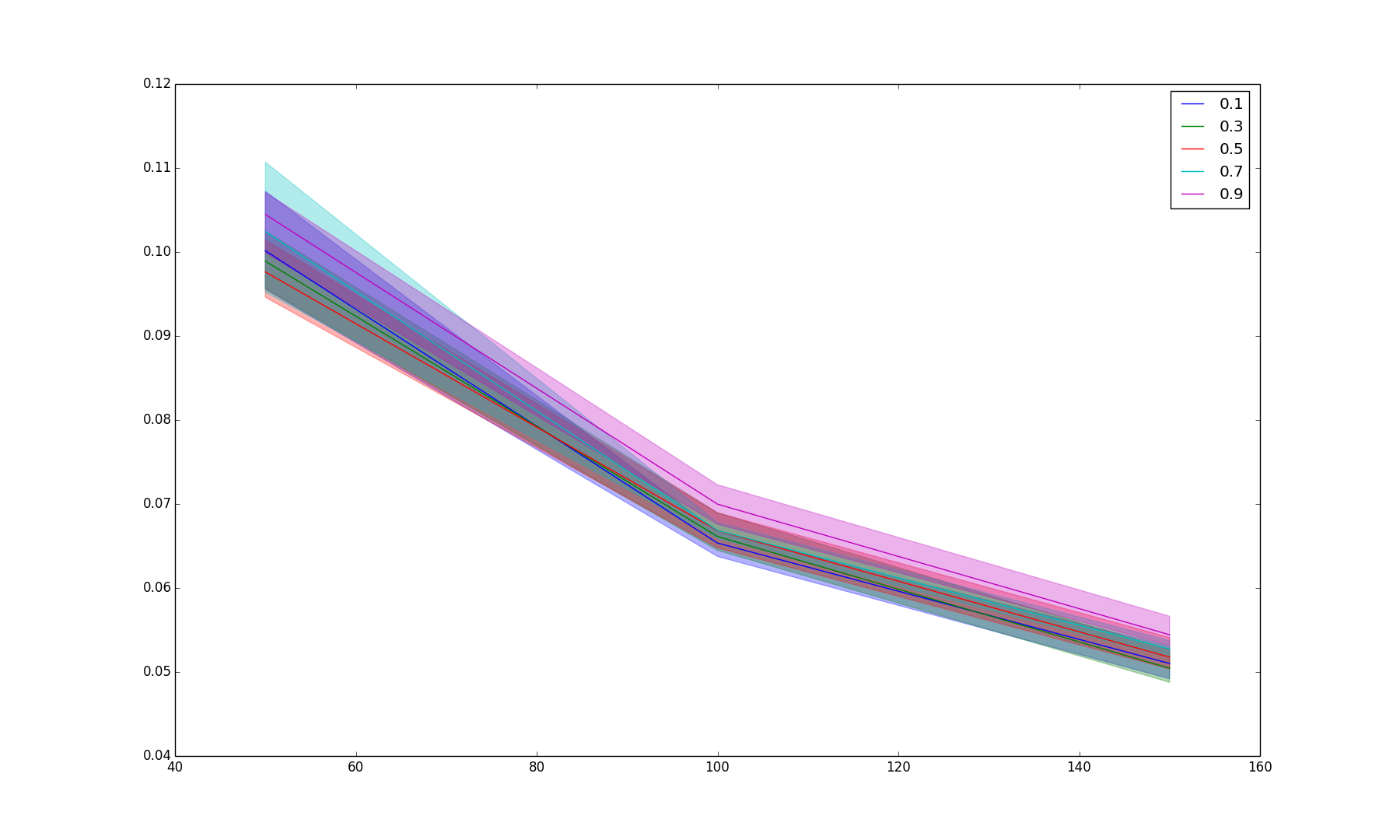
BIN
Results/res_temp_2_zoom.png
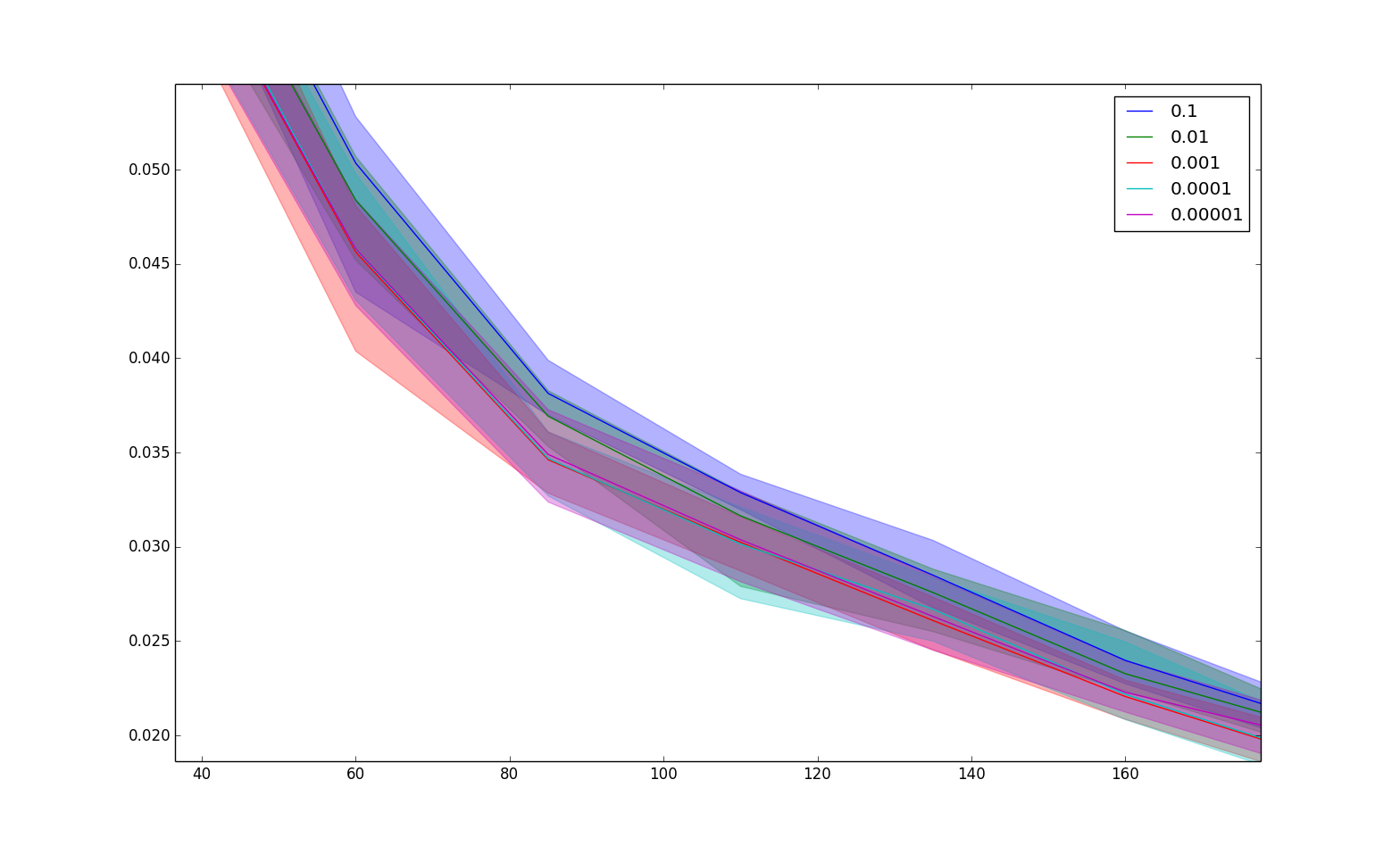
BIN
Results/resu_2_temp.png
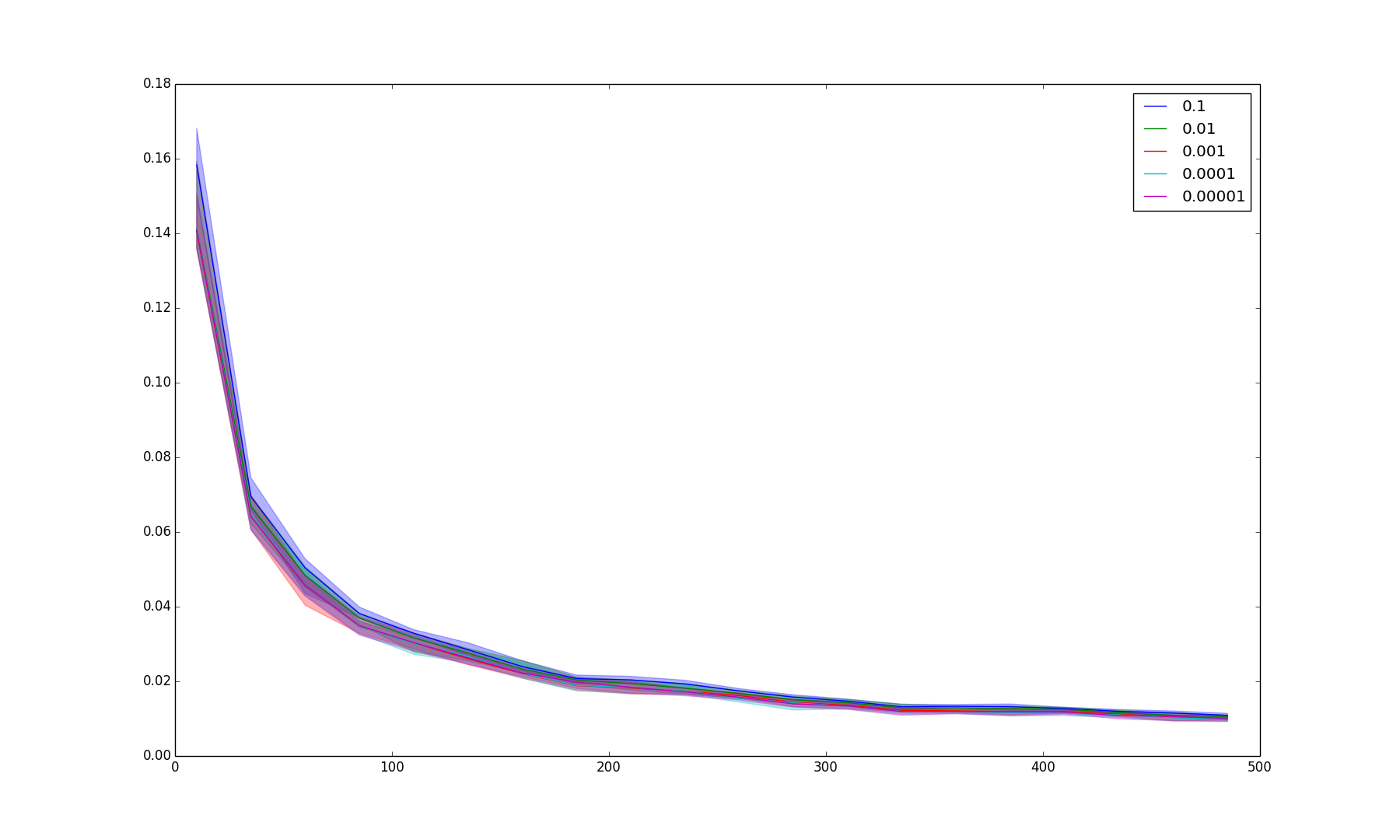
BIN
Results/resu_temp3.png
BIN
Results/resu_temp3_zoom.png

BIN
Results/sa_iter.png
BIN
Results/wrap_2.png
BIN
Results/wrap_2_zoom.png
BIN
Results/wrap_3.png
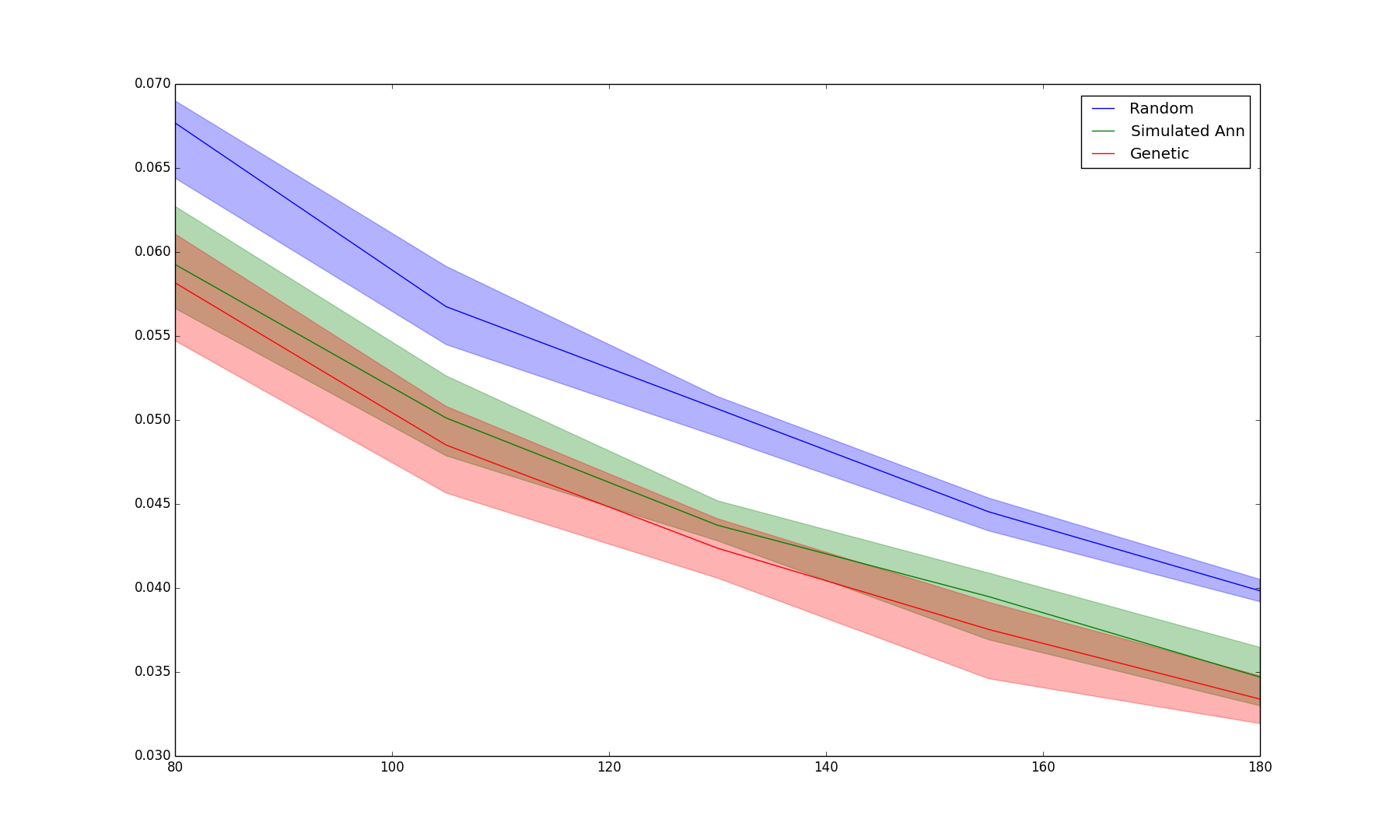
BIN
Results/wrap_4.png
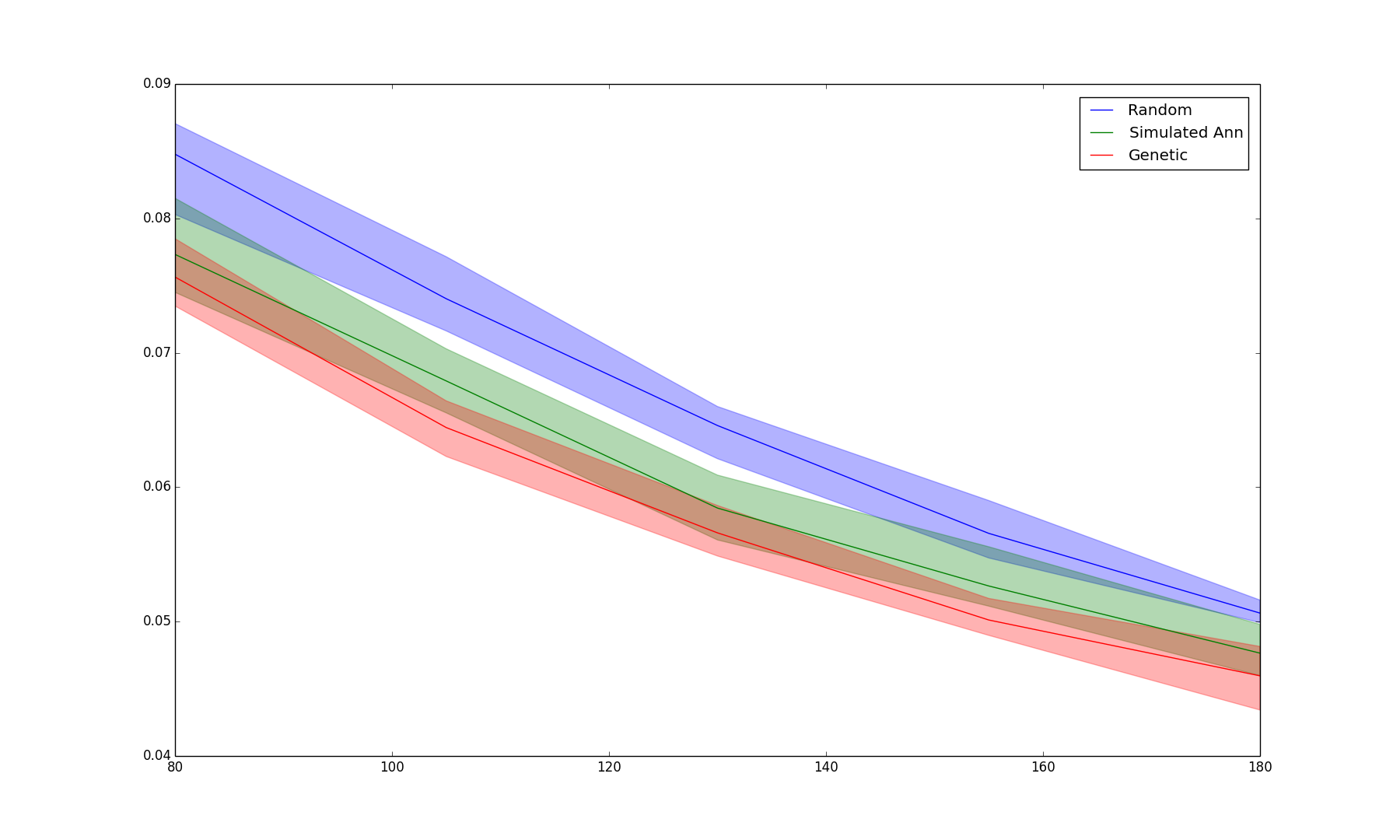
+ 26
- 4
main.tex
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 11
- 0
src/legend.py
|
|||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
|
|
||
+ 1
- 0
src/res_genetic_i22
|
|||
|
|
||